
dendrogram ( Z, leaf_label_func = llf, leaf_rotation = 90 ) # leaf_label_func can also be used together with ``truncate_mode`` parameter, # in which case you will get your leaves labeled after truncation: dendrogram ( Z, leaf_label_func = llf, leaf_rotation = 90, truncate_mode = 'level', p = 2 ) show_contracted bool, optional def llf ( id ): if id < n : return str ( id ) else : return '' % ( id, count, R ) # The text for the leaf nodes is going to be big so force # a rotation of 90 degrees. Non-singletons with their id, count, and inconsistency While indices \(k \geq n\) correspond to non-singletonįor example, to label singletons with their node id and Indices \(k < n\) correspond to original observations Is expected to return a string with the label for the When leaf_label_func is a callable function, for each leaf_label_func lambda or function, optional Unspecified, the size based on the number of nodes in theĭendrogram. Specifies the font size (in points) of the leaf labels. When unspecified, the rotation is based on the number of Specifies the angle (in degrees) to rotate the leaf When True, no labels appear next to the leaf nodes in the Useful if only the data structures computed for the renderingĪre needed or if matplotlib is not available. When True, the final rendering is not performed. Observation are labeled with the number of observations theyĬontain in parentheses. J appears in position i in the left-to-right traversal Includes a list R=H in the resultĭictionary. Than or equal to zero, all nodes are colored 'C0'.Ĭorresponding with MATLAB(TM) behavior, the threshold is set toĠ.7*max(Z).
All links connecting nodes withĭistances greater than or equal to the threshold are colored \(k\) the same color if \(k\) is the first node below color_threshold double, optionalįor brevity, let \(t\) be the color_threshold.Ĭolors all the descendent links below a cluster node Note: 'mtica' is an alias for 'level' that’s kept forīackward compatibility. No more than p levels of the dendrogram tree are displayed.Ī “level” includes all nodes with p merges from the final merge. Only non-leaf nodes in the linkage they correspond to rows The last p non-singleton clusters formed in the linkage are the Note: 'none' is an alias for None that’s kept for Truncation is used to condense the dendrogram. Observation matrix from which the linkage is derived is The dendrogram can be hard to read when the original
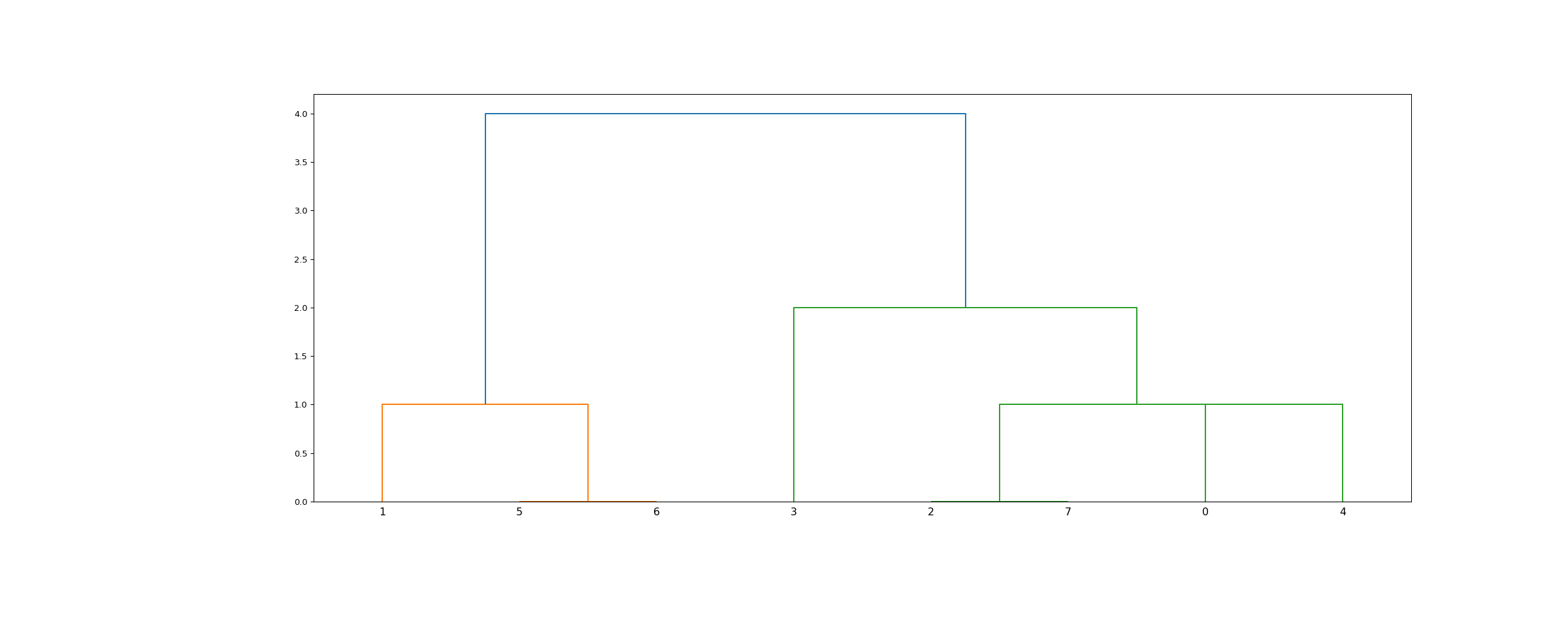
The linkage matrix encoding the hierarchical clustering to It is also theĬophenetic distance between original observations in the two The length of the two legs of the U-link represents The two legs of the U-link indicate which clusters The top of the U-link indicates aĬluster merge. The dendrogram illustrates how each cluster isĬomposed by drawing a U-shaped link between a non-singletonĬluster and its children. Plot the hierarchical clustering as a dendrogram. dendrogram ( Z, p = 30, truncate_mode = None, color_threshold = None, get_leaves = True, orientation = 'top', labels = None, count_sort = False, distance_sort = False, show_leaf_counts = True, no_plot = False, no_labels = False, leaf_font_size = None, leaf_rotation = None, leaf_label_func = None, show_contracted = False, link_color_func = None, ax = None, above_threshold_color = 'C0' ) ¶ Statistical functions for masked arrays ( K-means clustering and vector quantization (


 0 kommentar(er)
0 kommentar(er)
